Basalt¶
XML Input File¶
The basename for this file is basalt_circle.xml.
The file can be run using this command:
microstructpy --demo=basalt_circle.xml
The full text of the file is:
<?xml version="1.0" encoding="UTF-8"?>
<input>
<material>
<name> Plagioclase </name>
<fraction>
<dist_type> norm </dist_type>
<loc> 45.2 </loc>
<scale> 0.2 </scale>
</fraction>
<size>
<dist_type> cdf </dist_type>
<filename> aphanitic_cdf.csv </filename>
</size>
<color> #BDBDBD </color>
</material>
<material>
<name> Olivine </name>
<shape> ellipse </shape>
<fraction>
<dist_type> norm </dist_type>
<loc> 19.1 </loc>
<scale> 0.2 </scale>
</fraction>
<size>
<dist_type> cdf </dist_type>
<filename> aphanitic_cdf.csv </filename>
</size>
<aspect_ratio>
<dist_type> uniform </dist_type>
<loc> 1.0 </loc>
<scale> 2.0 </scale>
</aspect_ratio>
<angle_deg>
<dist_type> uniform </dist_type>
<loc> -90 </loc>
<scale> 180 </scale>
</angle_deg>
<color> #99BA73 </color>
</material>
<material>
<name> Diopside </name>
<fraction>
<dist_type> norm </dist_type>
<loc> 13.2 </loc>
<scale> 0.2 </scale>
</fraction>
<size>
<dist_type> cdf </dist_type>
<filename> aphanitic_cdf.csv </filename>
</size>
<color> #709642 </color>
</material>
<material>
<name> Hypersthene </name>
<fraction>
<dist_type> norm </dist_type>
<loc> 16.6 </loc>
<scale> 0.2 </scale>
</fraction>
<size>
<dist_type> cdf </dist_type>
<filename> aphanitic_cdf.csv </filename>
</size>
<color> #876E59 </color>
</material>
<material>
<name> Magnetite </name>
<fraction>
<dist_type> norm </dist_type>
<loc> 3.35 </loc>
<scale> 0.2 </scale>
</fraction>
<size>
<dist_type> cdf </dist_type>
<filename> aphanitic_cdf.csv </filename>
</size>
<color> #6E6E6E </color>
</material>
<material>
<name> Chromite </name>
<fraction>
<dist_type> norm </dist_type>
<loc> 0.65 </loc>
<scale> 0.2 </scale>
</fraction>
<size>
<dist_type> cdf </dist_type>
<filename> aphanitic_cdf.csv </filename>\
</size>
<color> #545454 </color>
</material>
<material>
<name> Ilmenite </name>
<fraction>
<dist_type> norm </dist_type>
<loc> 0.65 </loc>
<scale> 0.2 </scale>
</fraction>
<size>
<dist_type> cdf </dist_type>
<filename> aphanitic_cdf.csv </filename>
</size>
<color> #6B6B6B </color>
</material>
<material>
<name> Apatite </name>
<fraction>
<dist_type> norm </dist_type>
<loc> 3.35 </loc>
<scale> 0.2 </scale>
</fraction>
<size>
<dist_type> cdf </dist_type>
<filename> aphanitic_cdf.csv </filename>
</size>
<color> #ABA687 </color>
</material>
<domain>
<shape> circle </shape>
<diameter> 6 </diameter>
</domain>
<settings>
<directory> basalt_circle </directory>
<filetypes>
<seeds_plot> png </seeds_plot>
<poly_plot> png </poly_plot>
<tri_plot> png </tri_plot>
<verify_plot> png </verify_plot>
</filetypes>
<plot_axes> False </plot_axes>
<verbose> True </verbose>
<verify> True </verify>
<mesh_max_edge_length> 0.01 </mesh_max_edge_length>
<mesh_min_angle> 20 </mesh_min_angle>
<mesh_max_volume> 0.05 </mesh_max_volume>
<seeds_kwargs>
<linewidths> 0.2 </linewidths>
</seeds_kwargs>
<poly_kwargs>
<linewidths> 0.2 </linewidths>
</poly_kwargs>
<tri_kwargs>
<linewidths> 0.1 </linewidths>
</tri_kwargs>
</settings>
</input>
Material 1 - Plagioclase¶
Plagioclase composes approximately 45% of this basalt sample. It is an aphanitic component, meaning fine-grained, and follows a custom size distribution.
Material 2 - Olivine¶
Olivine composes approximately 19% of this basalt sample and is also an aphanitic component. The orientation of the olivine crystals is uniform random, with the aspect ratio varying from 1 to 3 uniformly.
Materials 3-8¶
Diopside, hypersthene, magnetite, chromite, ilmenite, and apatie compose approximately 36% of this basalt sample. They are aphanitic components and follow a custom size distribution, but are generally circular in shape.
Domain Geometry¶
These materials fill a circular domain with a diameter of 6 mm.
Settings¶
The function will output plots of the microstructure process and those plots
are saved as PNGs.
They are saved in a folder named basalt_circle, in the current directory
(i.e ./basalt_circle).
The axes are turned off in these plots, creating PNG files with minimal whitespace.
Mesh controls are introducted to increase grid resolution, particularly at the grain boundaries.
Output Files¶
The three plots that this file generates are the seeding, the polygon mesh, and the triangular mesh. These three plots are shown in Fig. 10 - Fig. 12.
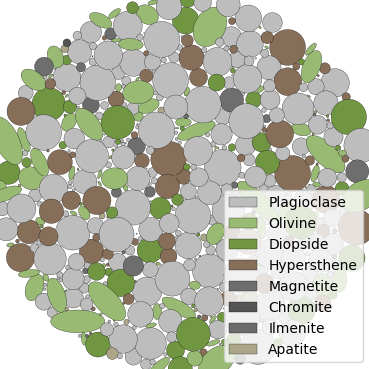
Fig. 10 Basalt example - seed geometries¶
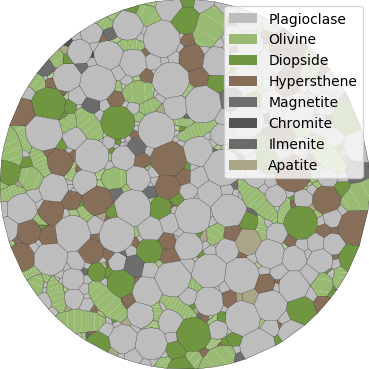
Fig. 11 Basalt example - polygonal mesh¶
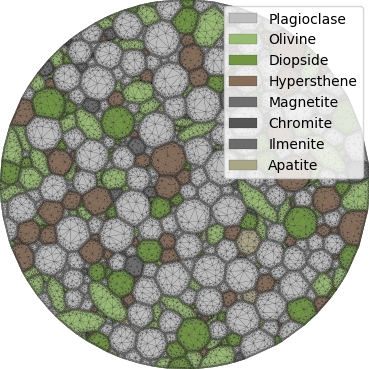
Fig. 12 Basalt example - triangular mesh¶
With the <verification> flag set to True, verification plots are
generated by MicroStructPy.
The grain size distribution comparison is given in Fig. 13.
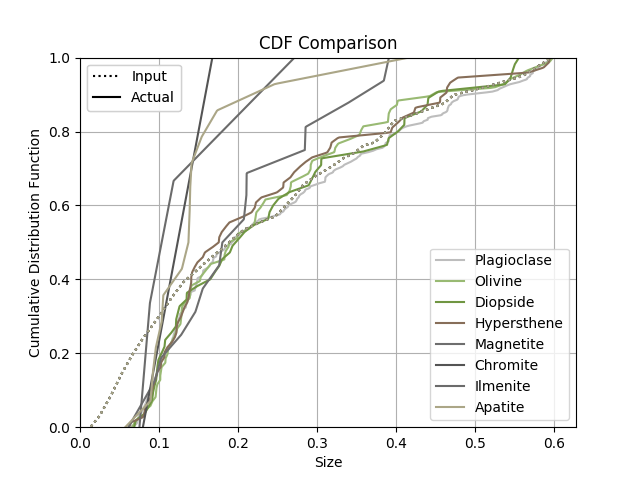
Fig. 13 Basalt example - input and output crystal size distributions (CSDs).¶
Comparing the input and output distributions, it is clear that this microstructure is not statistically representative. A larger diameter for the domain would contain more grains of olivine, which would add more fidelity to the size CDF curve.