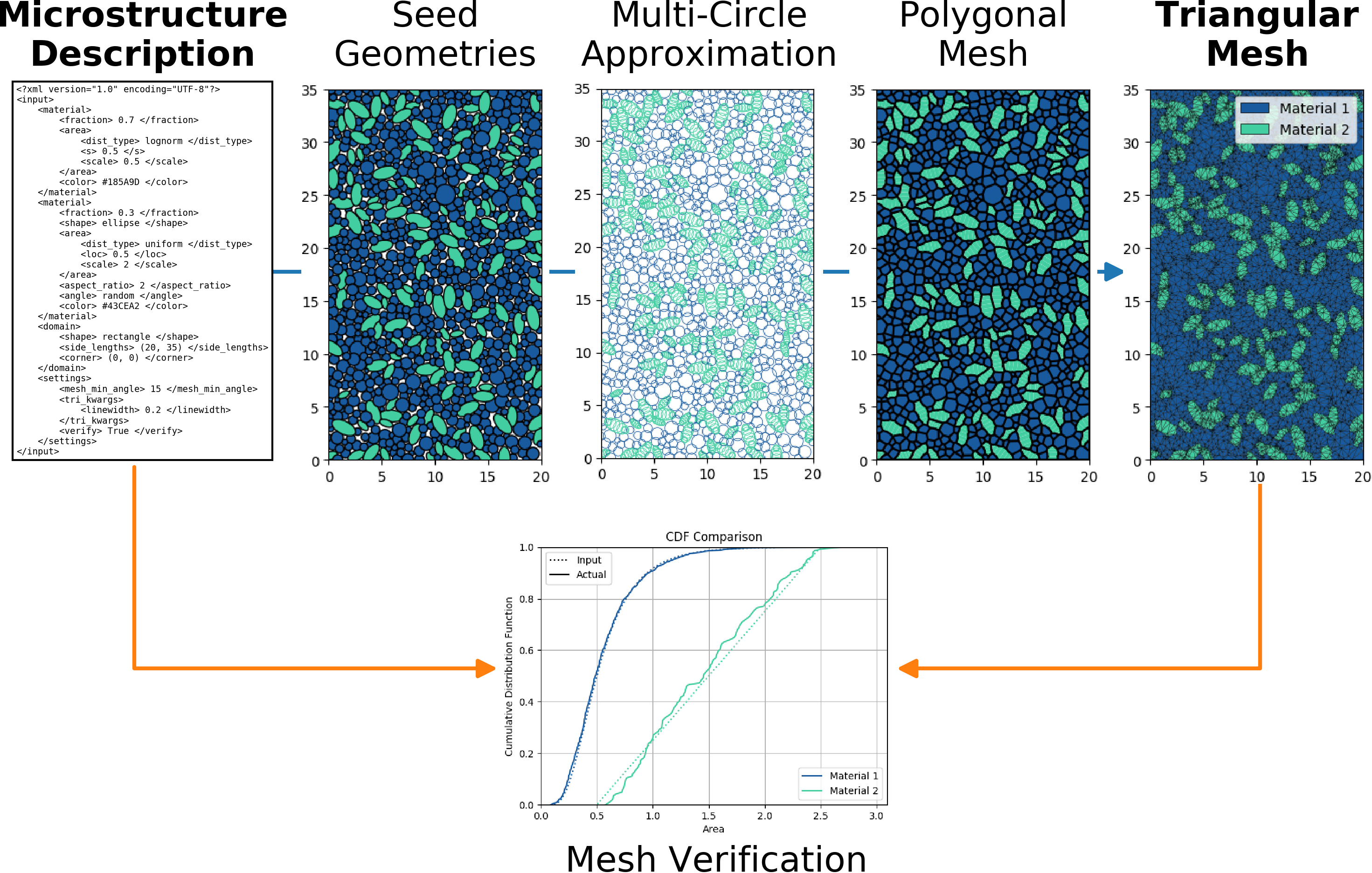
MicroStructPy Welcome Flowchart¶
Python Script¶
The basename for this file is msp_process.py.
The file can be run using this command:
microstructpy --demo=msp_process.py
The full text of the file is:
from __future__ import division
import os
import matplotlib.pyplot as plt
import microstructpy as msp
import numpy as np
from matplotlib.offsetbox import OffsetImage, TextArea, AnnotationBbox
inp_str = """
<?xml version="1.0" encoding="UTF-8"?>
<input>
<material>
<fraction> 0.7 </fraction>
<area>
<dist_type> lognorm </dist_type>
<s> 0.5 </s>
<scale> 0.5 </scale>
</area>
<color> #185A9D </color>
</material>
<material>
<fraction> 0.3 </fraction>
<shape> ellipse </shape>
<area>
<dist_type> uniform </dist_type>
<loc> 0.5 </loc>
<scale> 2 </scale>
</area>
<aspect_ratio> 2 </aspect_ratio>
<angle> random </angle>
<color> #43CEA2 </color>
</material>
<domain>
<shape> rectangle </shape>
<side_lengths> (20, 35) </side_lengths>
<corner> (0, 0) </corner>
</domain>
<settings>
<mesh_min_angle> 15 </mesh_min_angle>
<tri_kwargs>
<linewidth> 0.2 </linewidth>
</tri_kwargs>
<verify> True </verify>
</settings>
</input>
""".lstrip()
def main():
# Filenames
file_dir = os.path.dirname(os.path.realpath(__file__))
output_dir = os.path.join(file_dir, 'msp_process')
xml_basename = 'process.xml'
if not os.path.exists(output_dir):
os.makedirs(output_dir)
# Write XML File
xml_filename = os.path.join(output_dir, xml_basename)
with open(xml_filename, 'w') as file:
file.write(inp_str)
# Run XML File
in_data = msp.cli.read_input(xml_filename)
phases = in_data['material']
domain = in_data['domain']
kwargs = in_data['settings']
msp.cli.run(phases, domain, **kwargs)
# Plot Seed Breakdowns
seedlist_filename = os.path.join(output_dir, 'seeds.txt')
seeds = msp.seeding.SeedList.from_file(seedlist_filename)
plot_breakdown(seeds, phases, domain, output_dir)
# Combine Plots
zoom_nom = 0.086
px_nom = 742
w = 0.78
fig = plt.figure()
ax = fig.add_axes([0, 0, w, w])
ax.set_xlim(-11, 40)
ax.set_ylim(0.5, 10)
ax.set_axis_off()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# Arrow
y_xml_bot = 5.6
y_top = 9
y_mid = 0.5 * (y_xml_bot + y_top)
x_min = -6.1
x_side = 30.4
x_max = 35
x_mid = 0.5 * (x_min + x_max)
x_verif_r = x_mid + 7
y_verif_mid = 3.7
x_verif_l = x_mid - 7
cstyle = "angle,angleA=180,angleB=-90,rad=0"
ax.annotate("",
xy=(x_side, y_mid), xycoords='data',
xytext=(x_min, y_mid), textcoords='data',
arrowprops=dict(arrowstyle='-|>', color="C0",
patchA=None, patchB=None
),
)
ax.annotate("",
xy=(x_max, y_xml_bot), xycoords='data',
xytext=(x_verif_r, y_verif_mid), textcoords='data',
arrowprops=dict(arrowstyle='<|-', color="C1",
patchA=None, patchB=None,
connectionstyle=cstyle,
),
)
ax.annotate("",
xy=(x_min, y_xml_bot), xycoords='data',
xytext=(x_verif_l, y_verif_mid), textcoords='data',
arrowprops=dict(arrowstyle='<|-', color="C1",
patchA=None, patchB=None,
connectionstyle=cstyle,
),
)
# - Text of XML file
ob_xml = TextArea(inp_str.rstrip(),
textprops={'fontsize': 2.3, 'family': 'monospace'})
ab_xml = AnnotationBbox(ob_xml, (-10.4, 9 - 0.01), pad=0.1,
box_alignment=(0, 1),
bboxprops={'linewidth': 0.5})
ax.add_artist(ab_xml)
# - Seeds Plot
arr_seeds = plt.imread(os.path.join(output_dir, 'seeds.png'))
zoom = zoom_nom * px_nom / arr_seeds.shape[1]
ob_seeds = OffsetImage(arr_seeds, zoom=zoom)
ab_seeds = AnnotationBbox(ob_seeds, (0, 9),
pad=0, box_alignment=(0, 1),
bboxprops={'edgecolor': 'none'})
ax.add_artist(ab_seeds)
# - Breakdown Plot
bkdwn_filename = os.path.join(output_dir, 'breakdown.png')
remove_whitespace(bkdwn_filename)
arr_bkdwn = plt.imread(bkdwn_filename)
zoom = zoom_nom * px_nom / arr_bkdwn.shape[1]
ob_bkdwn = OffsetImage(arr_bkdwn, zoom=zoom)
ab_bkdwn = AnnotationBbox(ob_bkdwn, (10, 9),
pad=0, box_alignment=(0, 1),
bboxprops={'edgecolor': 'none'})
ax.add_artist(ab_bkdwn)
# - Polymesh Plot
arr_poly = plt.imread(os.path.join(output_dir, 'polymesh.png'))
zoom = zoom_nom * px_nom / arr_poly.shape[1]
ob_poly = OffsetImage(arr_poly, zoom=zoom)
ab_poly = AnnotationBbox(ob_poly, (20, 9),
pad=0, box_alignment=(0, 1),
bboxprops={'edgecolor': 'none'})
ax.add_artist(ab_poly)
# - Trimesh Plot
arr_tri = plt.imread(os.path.join(output_dir, 'trimesh.png'))
zoom = zoom_nom * px_nom / arr_tri.shape[1]
ob_tri = OffsetImage(arr_tri, zoom=zoom)
ab_tri = AnnotationBbox(ob_tri, (30, 9),
pad=0, box_alignment=(0, 1),
bboxprops={'edgecolor': 'none'})
ax.add_artist(ab_tri)
# - Verification Plot
verif_x = x_mid
verif_filename = os.path.join(output_dir, 'verification', 'area_cdf.png')
remove_whitespace(verif_filename)
arr_verif = plt.imread(verif_filename)
zoom = zoom_nom * 0.7 * 1645 / arr_verif.shape[1]
ob_verif = OffsetImage(arr_verif, zoom=zoom)
ab_verif = AnnotationBbox(ob_verif, (verif_x, 2.2),
pad=0, box_alignment=(0.5, 0),
bboxprops={'edgecolor': 'none'})
ax.add_artist(ab_verif)
# Add Titles
y_txt = 9.1
off = -0.1
fs = 9
plt.text(x_min + 0.3, y_txt, 'Microstructure\nDescription',
fontsize=fs, weight='bold', va='bottom', ha='center')
plt.text(5 + off, y_txt, 'Seed\nGeometries', fontsize=fs,
va='bottom', ha='center')
plt.text(15 + off, y_txt, 'Multi-Circle\nApproximation', fontsize=fs,
va='bottom', ha='center')
plt.text(25 + off, y_txt, 'Polygonal\nMesh', fontsize=fs,
va='bottom', ha='center')
plt.text(35 + off, y_txt, 'Triangular\nMesh', fontsize=fs,
weight='bold', va='bottom', ha='center')
plt.text(verif_x + 0.7, 2.1, 'Mesh Verification', fontsize=fs,
va='top', ha='center')
# Save Figure
out_filename = os.path.join(output_dir, 'process.png')
plt.savefig(out_filename, dpi=600)
plt.clf()
# Remove Whitespace
remove_whitespace(out_filename)
def plot_breakdown(seeds, phases, domain, output_dir):
plt.clf()
seed_colors = [phases[s.phase]['color'] for s in seeds]
seeds.plot_breakdown(edgecolors=seed_colors, facecolor='none',
linewidth=0.5)
lims = domain.limits
plt.axis('square')
plt.xlim(lims[0])
plt.ylim(lims[1])
plt.savefig(os.path.join(output_dir, 'breakdown.png'))
def remove_whitespace(filename):
im = plt.imread(filename)
rgb = im[:, :, :-1]
mask = rgb.min(axis=-1) < 1
col_mask = np.any(mask, axis=0)
row_mask = np.any(mask, axis=1)
row_i = np.argmax(row_mask)
row_j = len(row_mask) - np.argmax(np.flip(row_mask)) + 2
col_i = np.argmax(col_mask)
col_j = len(col_mask) - np.argmax(np.flip(col_mask)) + 2
new_im = im[row_i:row_j, col_i:col_j]
plt.imsave(filename, new_im)
if __name__ == '__main__':
main()
XML File¶
This example first writes an XML file and calls the CLI to produce
the output plots.
This file is saved to msp_process/process.xml.
Breakdown Plot¶
MicroStructPy does no produce plots of seed breakdowns by default.
To include this plot in the flow chart, the SeedList.plot_breakdown()
method is called, then the plot is formatted and saved.