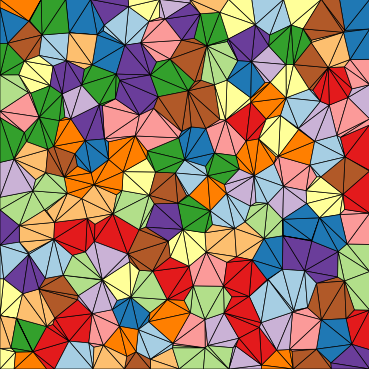
Minimal Example¶
XML Input File¶
The basename for this file is minimal_paired.xml.
The file can be run using this command:
microstructpy --demo=minimal_paired.xml
The full text of the file is:
<?xml version="1.0" encoding="UTF-8"?>
<input>
<material>
<shape> circle </shape>
<size> 0.09 </size>
</material>
<domain>
<shape> square </shape>
</domain>
<settings>
<directory> minimal </directory>
<filetypes>
<seeds_plot> png </seeds_plot>
<poly_plot> png </poly_plot>
<tri_plot> png </tri_plot>
</filetypes>
<plot_axes> False </plot_axes>
<color_by> seed number </color_by>
<colormap> Paired </colormap>
</settings>
</input>
Material 1¶
<material>
<shape> circle </shape>
<size> 0.09 </size>
</material>
There is only one material, with a constant size of 0.09.
Domain Geometry¶
<domain>
<shape> square </shape>
</domain>
The material fills a square domain. The default side length is 1, meaning the domain is greater than 10x larger than the grains.
Settings¶
<settings>
<directory> minimal </directory>
<filetypes>
<seeds_plot> png </seeds_plot>
<poly_plot> png </poly_plot>
<tri_plot> png </tri_plot>
</filetypes>
<plot_axes> False </plot_axes>
<color_by> seed number </color_by>
<colormap> Paired </colormap>
</settings>
The function will output plots of the microstructure process and those plots
are saved as PNGs.
They are saved in a folder named minimal, in the current directory
(i.e ./minimal).
The axes are turned off in these plots, creating PNG files with minimal whitespace.
Finally, the seeds and grains are colored by their seed number, not by material.